单细胞空间蛋白组分析技术服务
单细胞空间蛋白组分析是一项基于荧光标记抗体染色的超多标组织成像技术,该技术将多重蛋白表达谱的定性、定量信息与单细胞组织原位信息进行整合,可在一张病理切片上基于组织及细胞原位对多达60种蛋白进行多种分析。
非因生物建立的单细胞空间蛋白组分析平台为临床病理组织的空间细胞生物学和蛋白功能深度分析提供了全新的分析手段,平台拥有350+经严格验证的抗体资源库,支持结合研究方向和研究热点设计灵活的整体实验方案,是肿瘤微环境、免疫学肿瘤生物学、神经生物学、病理标志物诊断的全新研究利器。
技术原理
非因生物建立的单细胞空间蛋白组分析技术不同于常规多重免疫荧光技术(Multiplexed Immunofluorescence),是通过独特的多轮循环染色-成像-漂白流程,该流程可逆,且30轮后组织损伤<5%。
成像过程使用Cell DIVETM 自动化高清组织成像仪对5色荧光通道及明场通道进行扫描,并使用DAPI作为定位位点和内参、用HE图像对组织切片损伤度进行质控、用每轮采集的背景荧光图像做背景校正。搭载数据分析软件HALO来进行智能化的组织定量分析,可在完整组织图像单细胞水平对多个蛋白指标(含多种细胞类型标志物)进行精确空间定位和表达量分析,可真正实现对微环境内多种类型细胞准确鉴定,还可以对细胞密度和空间距离、免疫浸润、细胞间相互作用、细胞内部多种功能性蛋白之间的空间表达模式、相互关系以及与疾病的相关性进行深度解析。
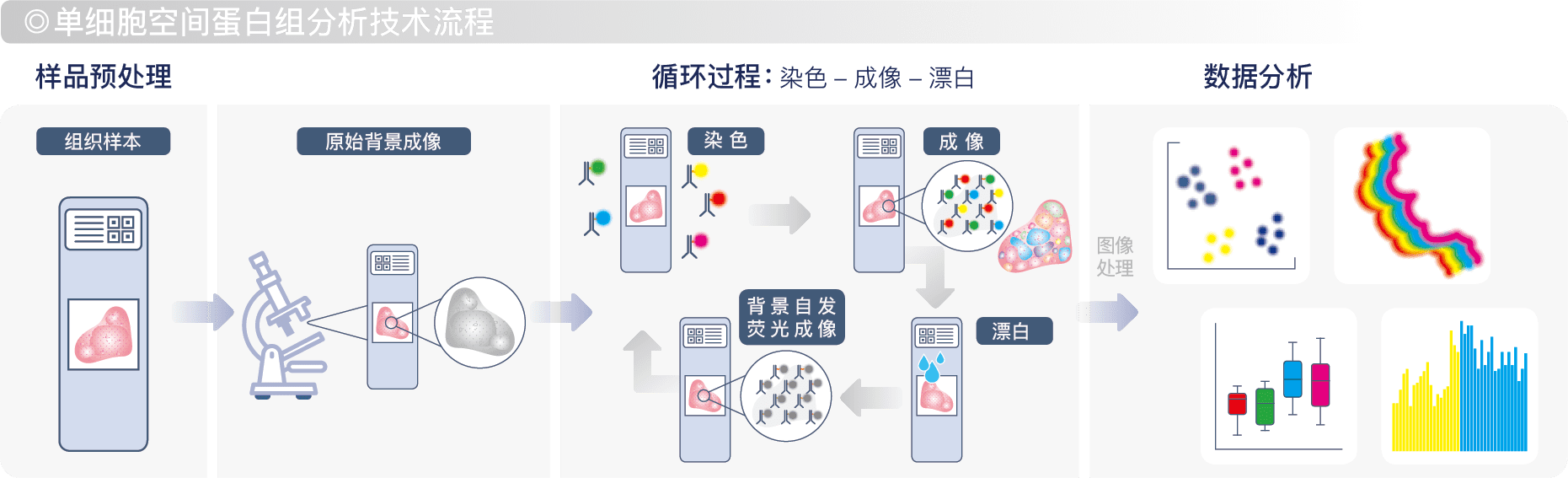
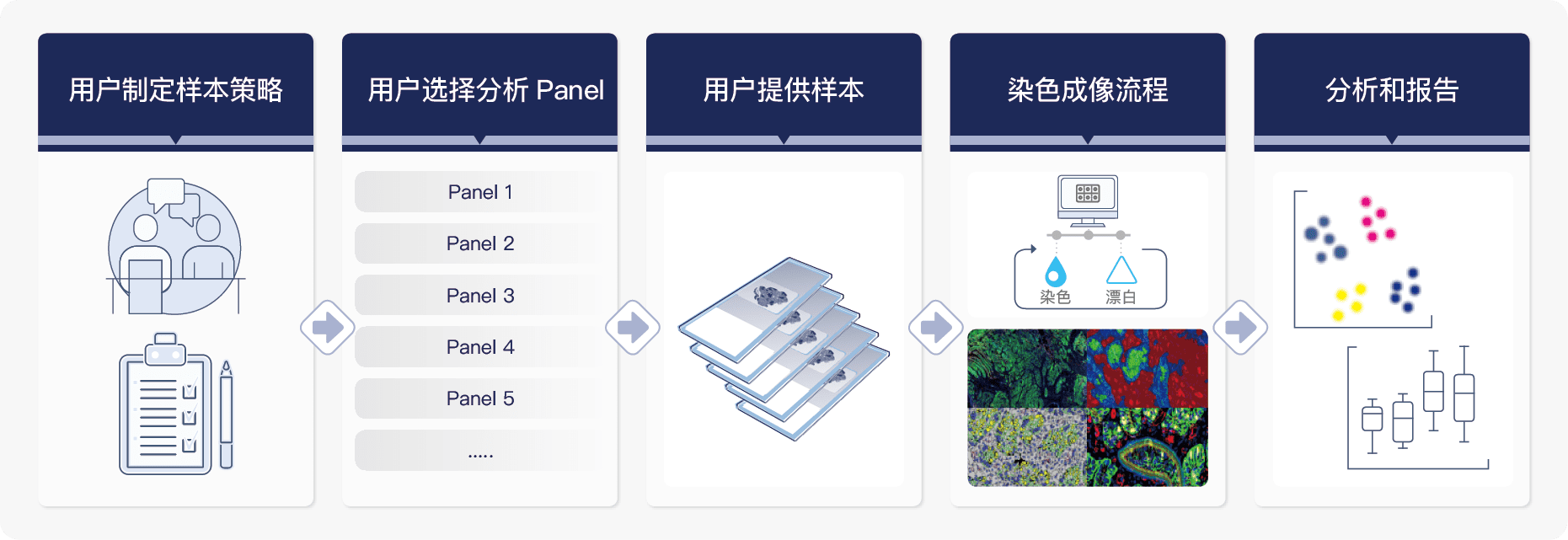
服务流程
靶点列表
肿瘤微环境分析
| CD69 | CD11b | CD57 | Cateninbeta | CD34 |
| CD11c | FOXP3 | CD33 | HLA-DR | CD31 |
| CD14 | CD163 | CD45RO | EGFR | CD68 |
| CD44 | PDL1 | CD8 | CD3 | CD4 |
| GFAP | SMA | CD20 | PD-1 | PanCK |
| E-cadherin | CD16(M) | SOX9 | Ki67 | CD45 |
| Vimentin | CD86 | CK8/18 | 持续增加中...... | |
TIL/TAM Panel – 10 markers
| TIL/TAM Panel | Co-expression | Phenotypes |
|---|---|---|
| CD3 | CD3+CD4+ | T helper |
| CD4 | CD3+CD4+FoxP3+ | T regulatory |
| CD8 | CD3+CD8+ | T cytotoxic |
| FoxP3 | CD68+ | TAM |
| CD68 | CD68+HLADR+CD163- | M1 TAM |
| CD163 | CD68+HLADR-CD163+ | M2 TAM |
| HLA-DR | CD34+ | Vessels |
| Ki67 | PanCK+Ki67+ | Proliferating tumor |
| CD34 | ||
| PanCK |
TIL Panel – 18 markers
| TIL panel | Co-expression | Phenotypes |
|---|---|---|
| CD3 | CD3+CD4+ | T helper |
| CD4 | CD3+CD4+FoxP3+ | T regulatory |
| CD8 | CD3+CD4+CD45RO+ | Memory T helper |
| FoxP3 | CD3+CD8+ | T cytotoxic |
| CD20 | CD3+CD8+CD45RO+ | Memory T cytotoxic |
| CD163 | CD68+HLADR+CD163- | M1 TAM |
| HLA-DR | CD68+HLADR-CD163+ | M2 TAM |
| CD57 | CD68+ | Macrophage |
| CD68 | CD20+ | B cell |
| PanCK | CD3-CD57+ | Natural Killer cell |
| SMA | CD11b+CD33+ | Myeloid cells |
| CD11b | CD11b+CD33+HLADR- | MDSC |
| CD33 | CD11b+HLADR-CD16+ | Neutrophil/G-MDSC |
| CD45RO | CD11b+CD14+ | Monocytes |
| CD14 | CD3+CD4+PD1+ | Immune modulation |
| CD16 | CD3+CD8+PD1+ | Immune modulation |
| PD-1 | CD68+PDL1+ | Macrophage PD-L1 |
| PD-L1 | CD20+PDL1+ | B cell PD-L1 |
TIL&Tumor Panel – 18 markers
| TIL&Tumor panel | Co-expression | Phenotypes |
|---|---|---|
| CD3 | CD3+CD4+ | T helper |
| CD4 | CD3+CD4+FoxP3+ | T regulatory |
| CD8 | CD3+CD8+ | T cytotoxic |
| FoxP3 | CD68+PDL1+ | Macrophage PD-L1 |
| CD20 | CD20+PDL1+ | B cell PD-L1 |
| CD163 | CD68+HLADR+CD163- | M1 TAM |
| HLA-DR | CD68+HLADR-CD163+ | M2 TAM |
| CD57 | CD68+ | Macrophage |
| CD68 | CD20+ | B cell |
| PanCK | CD3-CD57+ | Natural Killer cell |
| SMA | CD11b+CD33+ | Myeloid cells |
| CD11b | CD11b+CD33+HLADR- | MDSC |
| CD33 | CD11b+HLADR-CD16+ | Neutrophil/G-MDSC |
| PDL1 | CD11b+CD14+ | Monocytes |
| CD14 | Panck+KI67+ | Tumor cell KI67 |
| CD16 | Panck+EGFR+ | Tumor cell EGFR |
| KI67 | PanCK+PDL1+ | Tumor cell PD-L1 |
| EGFR |
Cell DIVE技术服务特点与优势
基于组织原位单细胞水平的高集成、高内涵分析
可在一张组织切片中,在完整组织图像单细胞水平对10重以上蛋白指标(含多种细胞类型标志物)进行精确空间定位和表达量分析,可真正实现对微环境内多种类型细胞准确鉴定,还可以对细胞密度和空间距离、免疫浸润、细胞间相互作用、细胞内部多种功能性蛋白之间的空间表达模式、相互关系以及与疾病的相关性进行深度解析。
灵活的实验设计方案
拥有350+个经严格验证的抗体资源库,根据实验需要灵活选择荧光抗体进行组合搭配;可根据研究需求自由选取样品的兴趣区域(ROI)进行后续多种分析。
严格验证的技术流程,精准的检测数据结果
经过非因生物严格验证、测试和优化的实验流程,无需剥离抗体或采取复杂的样本制备步骤,30轮实验过后,组织损伤仍然<5%;先进精准的仪器和自动化功能,可获取珍贵的实验数据,保证结果准确可靠。
强大的数据综合分析及深度挖掘能力
完全标准化的生物信息学分析流程,多年的空间组学相关数据分析经验,能够对生物学数据及临床数据进行综合分析及深度挖掘。
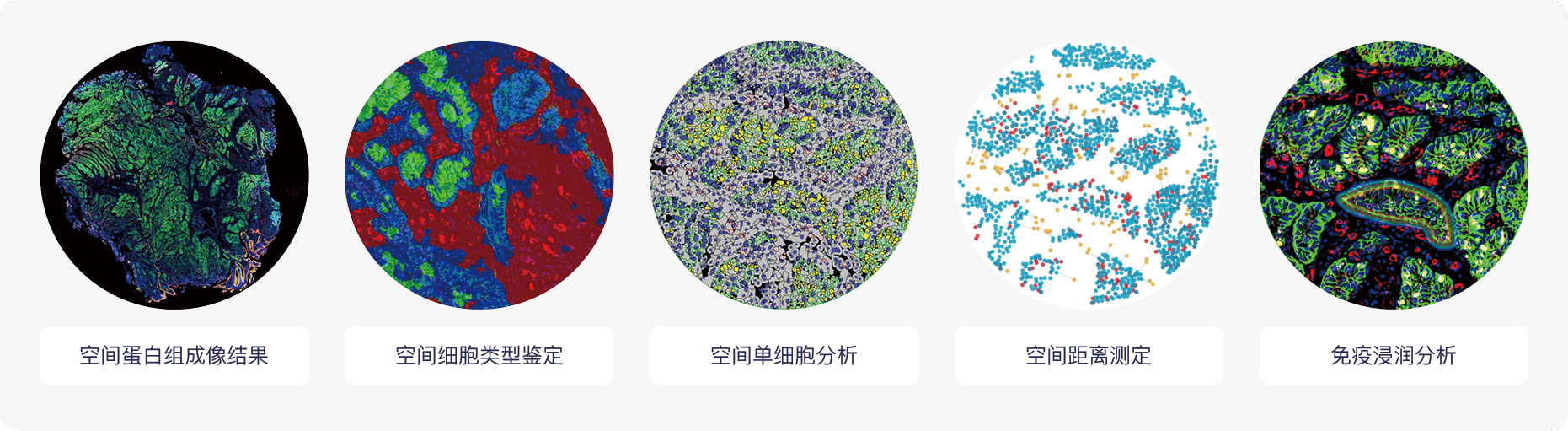
Cell DIVE技术服务的应用场景

示例结果:结直肠癌组织14种蛋白的成像与空间联合分析
参考文献
| PMID | 文献名 | 发表期刊 | 影响因子 |
|---|---|---|---|
| 28886381 | Oncolytic Virotherapy Promotes Intratumoral T Cell Infiltration and Improves Anti-PD-1 Immunotherapy | Cell | 66.85 |
| 24691442 | Excess PLAC8 promotes an unconventional ERK2-dependent EMT in colon cancer | The Journal of clinical investigation | 19.456 |
| 24463450 | Taxonomy of breast cancer based on normal cell phenotype predicts outcome | The Journal of clinical investigation | 19.456 |
| 31413142 | Regulatable interleukin-12 gene therapy in patients with recurrent high-grade glioma: Results of a phase 1 trial | Science translational medicine | 19.319 |
| 33771855 | Single-cell Spatial Proteomic Revelations on the Multiparametric MRI Heterogeneity of Clinically Significant Prostate Cancer | Clinical cancer research | 13.801 |
| 18559601 | The relative distribution of membranous and cytoplasmic met is a prognostic indicator in stage I and II colon cancer | Clinical cancer research | 13.801 |
| 33771855 | Single-cell Spatial Proteomic Revelations on the Multiparametric MRI Heterogeneity of Clinically Significant Prostate Cancer | Clinical cancer research | 13.801 |
| 34711609 | Heterogeneity of Circulating Tumor Cell-Associated Genomic Gains in Breast Cancer and Its Association with the Host Immune Response | Cancer Research | 13.312 |
| 29092944 | Platform for Quantitative Evaluation of Spatial Intratumoral Heterogeneity in Multiplexed Fluorescence Images | Cancer Research | 13.312 |
| 27197264 | Stromal-Based Signatures for the Classification of Gastric Cancer | Cancer research | 13.312 |
| 34711609 | Heterogeneity of Circulating Tumor Cell-Associated Genomic Gains in Breast Cancer and Its Association with the Host Immune Response | Cancer research | 13.312 |
| 26519361 | Cytometry-based single-cell analysis of intact epithelial signaling reveals MAPK activation divergent from TNF-α-induced apoptosis in vivo | Molecular systems biology | 13.068 |
| 31639039 | Efficacy and tolerability of anti-programmed death-ligand 1 (PD-L1) antibody (Avelumab) treatment in advanced thymoma | Journal for immunotherapy of cancer | 12.469 |
| 34599023 | Antitumor immune effects of preoperative sitravatinib and nivolumab in oral cavity cancer: SNOW window-of-opportunity study | J Immunother Cancer | 12.469 |
| 35013003 | Tumor MHC Class I Expression Associates with Intralesional IL2 Response in Melanoma | Cancer immunology research | 12.02 |
| 30745366 | Immune Profiling and Quantitative Analysis Decipher the Clinical Role of Immune-Checkpoint Expression in the Tumor Immune Microenvironment of DLBCL | Cancer immunology research | 12.02 |
| 29153838 | Unsupervised Trajectory Analysis of Single-Cell RNA-Seq and Imaging Data Reveals Alternative Tuft Cell Origins in the Gut | Cell systems | 11.091 |
| 34950863 | Characterization of the liver immune microenvironment in liver biopsies from patients with chronic HBV infection | JHEP Rep | 9.917 |
| 28570279 | Optimized multiplex immunofluorescence single-cell analysis reveals tuft cell heterogeneity | JCI Insight | 9.484 |
| 27182557 | Multiplexed immunofluorescence delineates proteomic cancer cell states associated with metabolism | JCI Insight | 9.484 |
| 33414541 | Multi-protein spatial signatures in ductal carcinoma in situ (DCIS) of breast | British journal of cancer | 9.075 |
| 34732839 | Stratification of chemotherapy-treated stage III colorectal cancer patients using multiplexed imaging and single-cell analysis of T-cell populations | Modern pathology | 8.209 |
| 34732839 | Stratification of chemotherapy-treated stage III colorectal cancer patients using multiplexed imaging and single-cell analysis of T-cell populations | Modern pathology | 8.209 |
| 29148540 | Single-cell heterogeneity in ductal carcinoma in situ of breast | Mod Pathol | 8.209 |
| 32616036 | Quantitative patterns of motor cortex proteinopathy across ALS genotypes | Acta neuropathologica communications. | 7.578 |
| 24854113 | A single slide multiplex assay for the evaluation of classical Hodgkin lymphoma | Am J Surg Pathol | 6.298 |
| 25566504 | Emerging understanding of multiscale tumor heterogeneity | Frontiers in oncology. | 5.738 |
| 29190747 | Characterizing the heterogeneity of tumor tissues from spatially resolved molecular measures | PLoS One | 3.752 |
| 31881020 | Multiscale, multimodal analysis of tumor heterogeneity in IDH1 mutant vs wild-type diffuse gliomas | Public Library of Science one | 3.752 |
| 31166985 | Understanding heterogeneous tumor microenvironment in metastatic melanoma | PLoS One | 3.752 |
| 24330149 | A novel, automated technology for multiplex biomarker imaging and application to breast cancer | Histopathology | 3.626 |
| 10054586 | Multi-modal imaging of histological tissue sections | IEEE | 3.557 |
| 16809241 | Robust single cell quantification of immune cell subtypes in histological samples | IEEE | 3.557 |
| Multi-channel algorithm for segmentation of tumor blood vessels using multiplexed image data | IEEE | 3.557 | |
| 27994939 | Pointwise mutual information quantifies intratumor heterogeneity in tissue sections labeled with multiple fluorescent biomarkers | Journal of pathology informatics | 3.23 |
| 27864340 | Microfluidic Tissue Mesodissection in Molecular Cancer Diagnostics | SLAS technology | 2.813 |
| 23818604 | Highly multiplexed single-cell analysis of formalin-fixed, paraffin-embedded cancer tissue | Proc Natl Acad Sci U S A |



















